The carnivorous bladderwort, a plant, has a tiny genome, only 80 million base pairs of DNA, six times fewer than a grape has, but it has more genes – 28,500 compared to the grape’s 26,300. Scientists believe they are able to explain what happened.
Biological Sciences Professor Victor Albert, who works at the University of Buffalo, described the bladderwort (Utricularia gibba) as a “marvel of nature”. It lives in water, has no discernible roots, has thread-like branches and tiny traps that use suction pressure to capture prey.
In this latest study, the authors published their findings and conclusions in the academic journal Molecular Biology and Evolution (citation below).
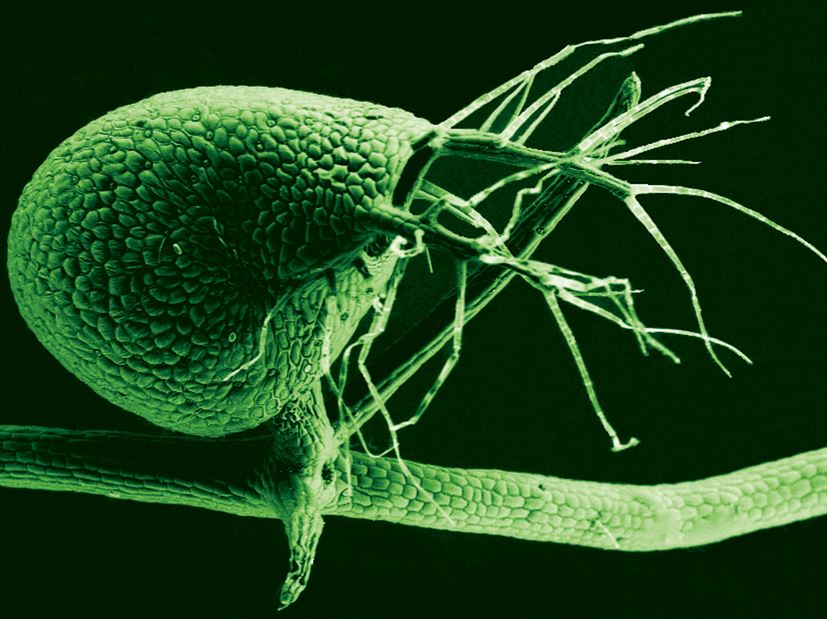
Scanning electron micrograph of the bladder of Utricularia gibba, the humped bladderwort plant. (Image: University of Buffalo)
The scientists found that the bladderwort has a smaller genome than a number of well-known plant species, such as coffee, papaya or grape, but has s much smaller genome.
Rampant DNA deletion
Study leader Prof. Albert explained that this amazing compact architecture resulted from a history of “rampant” DNA deletion in which the plant added and erased genetic material as an extremely fast pace.
The study also included partners from the Instituto de Ecología in Mexico, the Laboratorio Nacional de Genómica para la Biodiversidad (LANGEBIO) in Mexico, and the Universitat de Barcelona in Spain.
“The story is that we can see that throughout its history, the bladderwort has habitually gained and shed oodles of DNA.”
“With a shrunken genome,” he adds, “we might expect to see what I would call a minimal DNA complement: a plant that has relatively few genes — only the ones needed to make a simple plant. But that’s not what we see.”
Contrary to the minimalist plant theory, Prof. Albert and team found that U. gibba has more genes than several plants with larger genomes, including grape, as already noted, and Arabidopsis, a commonly-studied flower.
U. gibba is loaded with genes that facilitate carnivory – specifically, those that help it create enzymes similar to papain, a protein that helps break down meat fibers.
It is also rich in genes associated with the biosynthesis of cell walls, a vital feature of aquatic species that need to keep water at bay.
Prof. Albert said:
“When you have the kind of rampant DNA deletion that we see in the bladderwort, genes that are less important or redundant are easily lost. The genes that remain — and their functions — are the ones that were able to withstand this deletion pressure, so the selective advantage of having these genes must be pretty high.”
“Accordingly, we found a number of genetic enhancements, like the meat-dissolving enzymes, that make Utricularia distinct from other species.”
Most of the bladderowrt’s DNA that was deleted over time was “junk DNA” that contains no genes, the authors explained.
The team set out to determine how the bladderwort evolved the genetic structure it currently has. They compared it to four related species and discovered a pattern of rapid DNA alteration.
Prof. Albert explained:
“When you look at the bladderwort’s history, it’s shedding genes all the time, but it’s also gaining them at an appreciable enough rate, permitting it to stay alive and produce appropriate adaptations for its unique environmental niche.”
Replicated its entire genome
The researchers found that the U. gibba had undergone three duplication events which replicated its entire genome, giving it redundant copies of every gene.
This rapid gene gain was offset by fast-paced deletion. The fact that the plant has a tiny genome despite its history of genetic duplication is evidence of this phenomenon.
“In addition, the plant houses a high percentage of genes that don’t have close relatives within the genome, which suggests the plant quickly deleted redundant DNA acquired through duplication events,” the authors wrote.
Citation: “High gene family turnover rates and gene space adaptation in the compact genome of the carnivorous plant Utricularia gibba,” Lorenzo Carretero-Paulet, Pablo Librado, Tien-Hao Chang, Enrique Ibarra-Laclette, Luis Herrera-Estrella, Julio Rozas, and Victor A. Albert. Mol Biol Evol first published online January 31, 2015. DOI: 10.1093/molbev/msv020.

