The tree of life, developed by researchers at the University of California, Berkeley, shows the astonishing diversity of life on Earth – how life has evolved and spread in a wonderful and fascinating number of ways.
Jill Banfield, a UC Berkeley professor of earth and planetary science and environmental science, policy and management, and colleagues have discovered over 1,000 new types of bacteria and Archaea (a domain/kingdom of single-celled microorganisms) over the last fifteen years lurking in our planet’s nooks and crannies, and have dramatically redesigned the tree to account for these minuscule new life forms.
Prof. Banfield said:
“The tree of life is one of the most important organizing principles in biology. The new depiction will be of use not only to biologists who study microbial ecology, but also biochemists searching for novel genes and researchers studying evolution and earth history.”
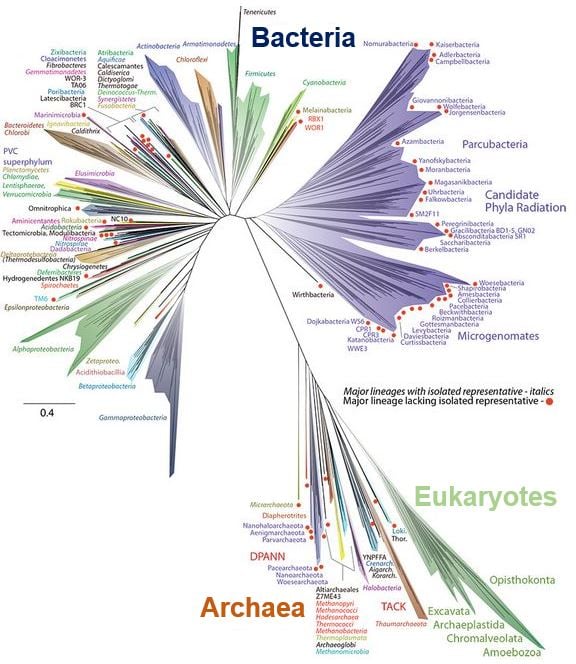 An artist’s representation of the Tree of Life, with the several groups of bacteria on the left, the uncultivable bacteria at upper right (purple), and the Archaea and eukaryotes (green) – which includes humans – at the lower right. (Image: news.berkeley.edu. Graphic by Zosia Rostomian, Lawrence Berkeley National Laboratory)
An artist’s representation of the Tree of Life, with the several groups of bacteria on the left, the uncultivable bacteria at upper right (purple), and the Archaea and eukaryotes (green) – which includes humans – at the lower right. (Image: news.berkeley.edu. Graphic by Zosia Rostomian, Lawrence Berkeley National Laboratory)
Genome revolution changed everything
A major part of this microbial diversity remained hidden until the genome revolution allowed scientists like Prof. Banfield to search directly for their genomes in various habitats, rather then attempting to culture them in a lab dish.
Many of the microbes cannot live on their own, so they cannot be cultured or isolated. They exist by begging, borrowing or stealing stuff from other microbes or animals, either as scavengers, symbiotic organisms or parasites.
The new tree has been published in the new journal Nature Microbiology (citation below). It reinforces once again that the life we see around us – animals, plants, humans and other so-called *eukaryotes – represent a tiny fraction of Earth’s biodiversity.
* Eukaryotes are organisms with a complex cell or cells, in which the genetic material is organized into a membrane-bound nucleus or nuclei. Animals, plants and fungi – which are mainly multicellular – are eukaryotes, as opposed to prokaryotes, such as Archaea and bacteria, which lack nuclei and other complex cell structures.
 Archaea are one-celled microorganisms genetically different from bacteria and commonly found in extreme environmental conditions, such as hot springs. They were first found in extreme environments. Pictured above is Grand Prismatic Spring of Yellowstone National Park. (Image: Wikipedia)
Archaea are one-celled microorganisms genetically different from bacteria and commonly found in extreme environmental conditions, such as hot springs. They were first found in extreme environments. Pictured above is Grand Prismatic Spring of Yellowstone National Park. (Image: Wikipedia)
Most of life’s diversity from bacteria and Archaea
Prof. Banfield, who also has an appointment at Lawrence Berkeley National Laboratory, said:
“Bacteria and Archaea from major lineages completely lacking isolated representatives comprise the majority of life’s diversity.”
“This is the first three-domain genome-based tree to incorporate these uncultivable organisms, and it reveals the vast scope of as yet little-known lineages.
Lead author, Dr. Laura Hug, who works at the biology faculty at the University of Waterloo, Ontario, Canada, and was a postdoctoral fellow at UC Berkeley, says that the more than 1,000 newly-reported organisms appearing on the updated tree are from a range of environments, including a salt flat in Chile’s Atacama desert, a hot spring in Yellowstone National Park, a sparkling water geyser, meadow soil, terrestrial and wetland sediments, and the inside of a dolphin’s mouth.
All these newly-discovered or newly-recognised organisms are known only from their genomes.
Dr. Hug said:
“What became really apparent on the tree is that so much of the diversity is coming from lineages for which we really only have genome sequences. We don’t have laboratory access to them, we have only their blueprints and their metabolic potential from their genome sequences.”
“This is telling, in terms of how we think about the diversity of life on Earth, and what we think we know about microbiology.”
 Charles Darwin (1809-1882) described the Tree of Life in a famous passage in his work ‘On the Origin of Species’, published in 1859. (Image: biography.com)
Charles Darwin (1809-1882) described the Tree of Life in a famous passage in his work ‘On the Origin of Species’, published in 1859. (Image: biography.com)
Candidate phyla radiation
One striking feature of the new tree of life is that a very major branch of it comprises a group of bacteria described as ‘candidate phyla radiation’. Only recently recognised and seemingly comprised exclusively of symbiotic lifestyles, the candidate phyla radiation appears to contain about half of all bacterial evolutionary diversity.
While the relationship between eukaryotes and Archaea remains unclear, it is evident that “this new rendering of the tree offers a new perspective on the history of life,” Prof. Banfield said.
Co-author, Assistant Professor Brett Baker, who works at the University of Texas’ Marine Science Institute, Austin, said:
“This incredible diversity means that there are a mind-boggling number of organisms that we are just beginning to explore the inner workings of that could change our understanding of biology.”
Tree portrays life we see today
The tree of life was first sketched in 1837 by Charles Darwin as he sought ways of showing how bacteria, animals and plants are related to one another. The idea gained traction in the 19th century, with the tips of twigs representing life on our planet today, while the branches connecting them to the trunk implied evolutionary associations among these creatures.
A branch that splits into two twigs near the tips of the tree suggests that these organisms have a recent ancestor in common, while a forking branch close to the trunk suggest an evolutionary diversion earlier on.
Archaea were first added to the tree in 1977, following work showing that they are distinctly different from bacteria, even though they are single-celled organisms like bacteria. Prof. Banfield said that a tree published in 1990 by microbiologist Carl Woese was “a transformative visualization of the tree.” With its three different domains, it is still the most recognisable today.
DNA sequencing sped up everything
As DNA sequencing became progressively easier since 2000, Prof. Banfield and others started sequencing whole communities of organisms simultaneously and picking out individual groups based just on their genes.
This metagenomic sequencing revealed entirely new groups of Archaea and bacteria, many of them living in extreme environments, such as the human gut, the dirt under toxic waste sites, and toxic puddles in abandoned mines.
Some of these had already been detected, but nobody knew anything about them because they could not be isolated in a lab dish (they died).
For the paper, Dr. Hug and Prof. Banfield teamed up with several scientists who have sequenced new microbial species, gathering 1,011 previously-unpublished genomes to add to already known genome sequences of organisms that represent the major families of life on Earth.
Prof. Banfield and her team built a tree based on sixteen separate genes that code for proteins in the cellular machine called a ribosome, a protein builder or the protein synthesizer of the cell, i.e. it translates RNA into proteins.
They included 3,083 organisms – one from each genus for which fully or nearly fully sequenced genomes are available.
The analysis, which represented the total diversity among all sequenced genomes, resulted in a tree with branches dominated by bacteria, especially by uncultivated bacteria.
A second view of the tree of life grouped organisms by their evolutionary distance from one another instead of current taxonomic definitions, clearly showing that around one-third of all biodiversity comes from bacteria, one-third from uncultivable bacteria and slightly less than one-third from eukaryotes and Archaea.
Proof. Banfield said:
“The two main take-home points I see in this tree are the prominence of major lineages that have no cultivable representatives, and the great diversity in the bacterial domain, most importantly, the prominence of candidate phyla radiation.”
“The candidate phyla radiation has as much diversity within it as the rest of the bacteria combined.”
In an Abstract in the journal, the authors wrote:
“The results reveal the dominance of bacterial diversification and underline the importance of organisms lacking isolated representatives, with substantial evolution concentrated in a major radiation of such organisms.”
“This tree highlights major lineages currently underrepresented in biogeochemical models and identifies radiations that are probably important for future evolutionary analyses.”
The study was supported mainly by the US Department of Energy through Lawrence Berkeley National Laboratory, with metagenomic sequencing by DOE’s Joint Genome Institute in Walnut Creek, California.
Citation: “A new view of the tree of life,” Jillian F. Banfield, Laura A. Hug, Brett J. Baker, Karthik Anantharaman, Ronald Amundson, Brian C. Thomas, Christopher T. Brown, Alexander J. Probst, Cindy J. Castelle, Cristina N. Butterfield, Alex W. Hernsdorf, Yuki Amano, Kotaro Ise, David A. Relman, Kari M. Finstad, Yohey Suzuki & Natasha Dudek. Nature Microbiology, Article number: 16048 (2016). DOI: 10.1038/nmicrobiol.2016.48

