Scientists say they have found the “Enigma Machine” that cracks the code of a whole class of viruses, including those that cause the common cold and polio. This means they probably have the means to eventually make the viruses harmless.
To date, researchers had not been aware of the code that had been hiding in plain sight in the sequence of the RNA (ribonucleic acid) that makes up this type of viral genome.
Scientists from the Universities of Leeds and York in England say they have managed to unlock the code and believe it is possible to jam it, thus disrupting virus assembly. If a virus cannot assemble, it will cease to function and not be able to cause disease.
The study has been published in the academic journal Proceedings of the National Academy of Sciences (citation below).
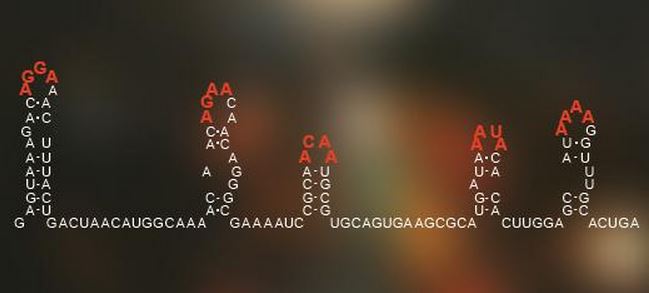
A code hidden in the arrangement of the genetic information of single-stranded RNA viruses tells it how to pack itself within its outer shell of proteins. (Image: University of Leeds)
Study leader, Professor Peter Stockley, who works in the Faculty of Biological Sciences at the University of Leeds, said:
“If you think of this as molecular warfare, these are the encrypted signals that allow a virus to deploy itself effectively.”
“Now, for this whole class of viruses, we have found the ‘Enigma machine’ – the coding system that was hiding these signals from us. We have shown that not only can we read these messages but we can jam them and stop the virus’ deployment.”
Single-stranded RNA viruses are simple, but many are potent
Single-stranded RNA viruses were probably the first ones to evolve; they are the simplest type of virus. Many of them are, however, some of the most powerful and damaging infectious pathogens.
Rhinovirus, the one that causes the common cold, causes about 1 billion infections every year globally, which is more than all the other infectious agents combined.
Other single-stranded RNA viruses include HIV, hepatitis C virus, norovirus (winter vomiting bug), and poliovirus.
This latest breakthrough was the result of three stages of research:
Stage 1: In 2012, University of Leeds scientists observed at a single-molecule level how the core of a single-stranded RNA virus packed itself into its outer shell. The core must first be correctly folded to fit into the protective viral protein coat.
The viruses solve this problem in fractions of a second. The researchers then set about the challenge of finding out how the viruses did this.
Stage 2: Dr. Eric Dykeman and Professor Reidun Twarock, two University of York mathematicians, worked together with the Leeds group and devised algorithms to crack the code governing the process. They then build computer-based models of the coding system.
Stage 3 (latest study): The two groups unlocked the code. They used single-molecule fluorescence spectroscopy to observe the codes being used by the satellite tobacco necrosis virus, a type of single-stranded RNA plant virus.
Dr. Roman Tuma, Reader in Biophysics at The Astbury Centre for Structural Molecular Biology, University of Leeds, said:
“We have understood for decades that the RNA carries the genetic messages that create viral proteins, but we didn’t know that, hidden within the stream of letters we use to denote the genetic information, is a second code governing virus assembly. It is like finding a secret message within an ordinary news report and then being able to crack the whole coding system behind it.”
“This paper goes further: it also demonstrates that we could design molecules to interfere with the code, making it uninterpretable and effectively stopping the virus in its tracks.”
Professor Twarock said:
“The Enigma machine metaphor is apt. The first observations pointed to the existence of some sort of a coding system, so we set about deciphering the cryptic patterns underpinning it using novel, purpose designed computational approaches.”
“We found multiple dispersed patterns working together in an incredibly intricate mechanism and we were eventually able to unpick those messages. We have now proved that those computer models work in real viral messages.”
The research team plans to experiment with animal viruses next. They believe their combination of single-molecule detection capabilities plus their computational models will offer a new route for drug discovery.
The study was sponsored by the BBSRC (Biotechnology and Biological Sciences Research Council), the EPSRC (Engineering and Physical Sciences Research Council), Dr Dykeman’s Leverhulme Trust Early Career Fellowship, and Professor Twarock’s Royal Society Leverhulme Trust Senior Research Fellowship.
Citation: “Revealing the density of encoded functions in a viral RNA,” Nikesh Patel, Eric C. Dykeman, Robert H. A. Coutts, George P. Lomonossoff, David J. Rowlands, Simon E. V. Phillips, Neil Ranson, Reidun Twarock, Roman Tuma, and Peter G. Stockley. PNAS 2015 ; published ahead of print February 2, 2015, doi:10.1073/pnas.1420812112.
